Welcome to TWILIGHT Wiki

TWILIGHT Video Tutorial
Introduction
Overview
TWILIGHT (Tall and Wide Alignments at High Throughput) is an innovative MSA tool designed to leverage massive parallelism for efficient handling of tall and wide alignment tasks. It is able to scale to millions of long nucleotide sequences (>10000 bases). TWILIGHT can run on CPU-only platforms (Linux/Mac) or take advantage of CUDA-capable GPUs for further acceleration.
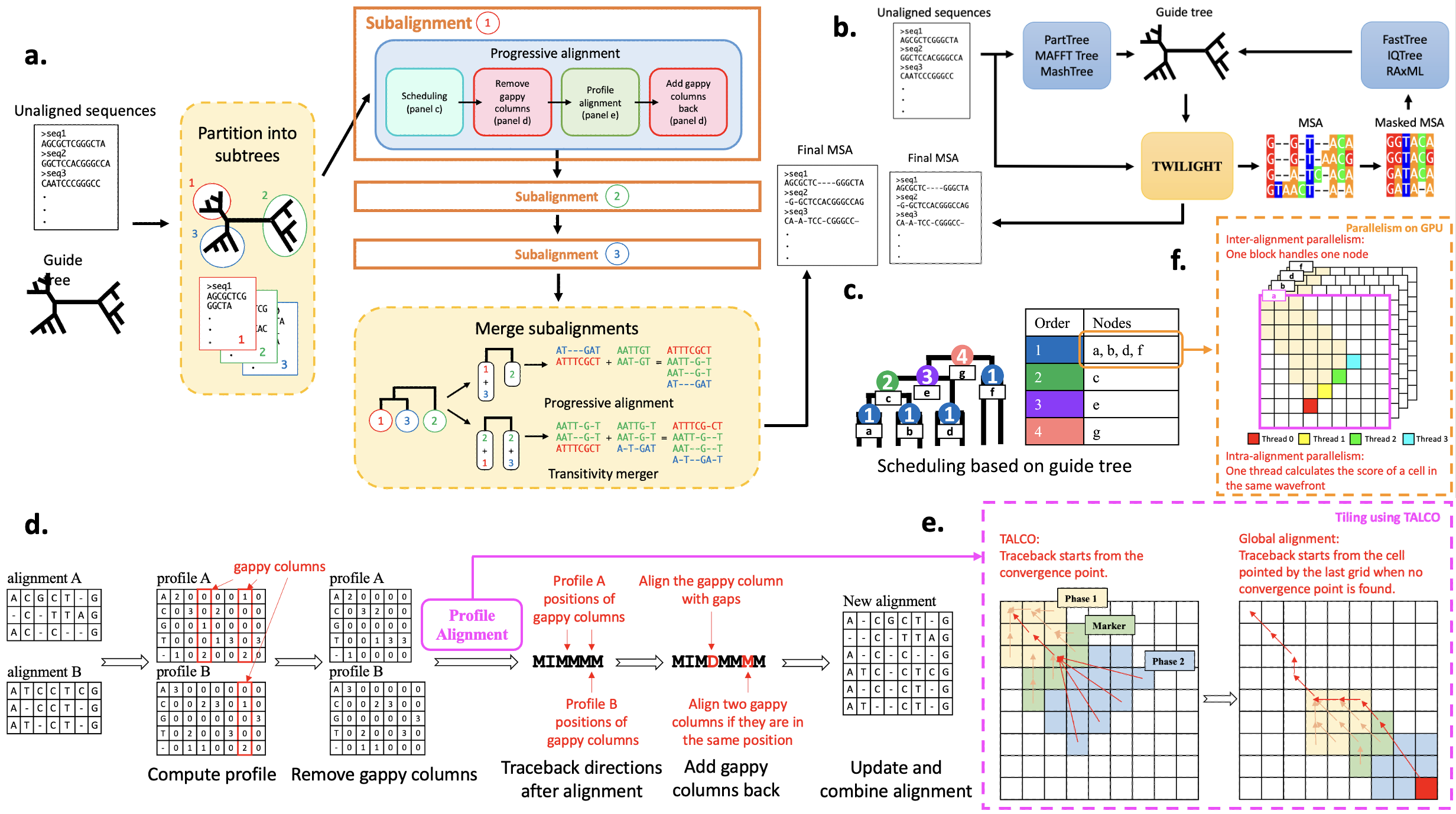
By default, TWILIGHT requires an unaligned sequence file in FASTA format and an input guide tree in Newick format to generate the output alignment in FASTA format (Fig. 1a, see TWILIGHT Default Mode for more details). When a guide tree is unavailable, users can utilize the iterative mode, which provides a Snakemake workflow to estimate guide trees using external tools (Fig. 1b, see TWILIGHT Iterative Mode for more details).
TWILIGHT adopts the progressive alignment algorithm (Fig. 1c) and employs tiling strategies to band alignments (Fig. 1e). Combined with a divide-and-conquer technique (Fig. 1a), a novel heuristic dealing with gappy columns (Fig. 1d) and support for GPU acceleration (Fig. 1f), TWILIGHT demonstrates exceptional speed and memory efficiency.
Key Features
Divide-and-Conquer Technique
TWILIGHT's divide-and-conquer technique (Fig. 1a) starts by dividing the initial guide tree into smaller subtrees with at most m leaves, which controls memory usage by limiting the number of sequences processed at once. TWILIGHT sequentially iterates over each subtree, loading its corresponding sequences into memory, and computing the subalignment for the subtree. After all subalignments are computed, TWILIGHT merges subalignments with transitivity merger algorithm of PASTA (Mirarab et al., 2015) or progressive alignment.
Progressive Alignment and Scheduling
TWILIGHT performs progressive alignment based on the guide tree structure, first aligning the most similar sequences or sequence groups and then progressively adding less similar sequences or groups. The scheduling algorithm (Fig. 1c) initializes the leaf nodes and then performs a post-order traversal to set the alignment order for internal nodes. This approach allows certain child nodes (e.g., a, b, d, f) to be aligned in parallel, while others (e.g., c, e, g) are processed sequentially, ensuring efficient parallel processing while respecting tree dependencies.
Removing gappy columns
As more sequences are added, the alignment tends to grow longer, often with many gap-dominated columns, leading to increased computational time. To improve efficiency, TWILIGHT excludes gappy columns (Fig. 1d)—where gaps exceed a specified threshold—before the alignment step, resulting in shorter profiles and reducing computational overhead. After the alignment step, TWILIGHT restores the excluded gappy columns. If gappy columns from both profiles align at the same position, they are merged, reducing the overall alignment length. Otherwise, a gappy column from one profile is aligned with a new gap introduced in the other profile.
Banded Alignment and Tiling Strategy
TWILIGHT employs a modified X-Drop algorithm to control memory usage in pairwise profile alignment, ensuring linear scaling with sequence length. However, storing traceback pointers in limited GPU shared memory still poses challenges. To address this, TWILIGHT integrates the TALCO tiling algorithm (Walia et al., 2024) (Fig. 1e), which limits traceback storage to a constant memory usage by using convergence pointers.
Parallelization Techniques
TWILIGHT employs parallelization techniques on both CPU and GPU to maximize efficiency. On CPUs, it is implemented in C++ with Threading Building Blocks (TBB) to exploit thread-level parallelism, processing independent alignments in parallel while also parallelizing profile calculations and sequence updates. On GPUs, TWILIGHT utilizes three levels of parallelism (Fig. 1f): multi-GPU parallelism distributes alignment tasks into batches and sends to multiple GPUs, inter-alignment parallelism assigns each GPU thread block to a pairwise profile alignment, and intra-alignment parallelism processes multiple cells in the dynamic programming matrix wavefront simultaneously.
Installation Methods
Installation summary (choose your installation method)
TWILIGHT offers multiple installation methods for different platforms and hardware setups:
- Conda is recommended for most users needing the default mode and partial iterative mode support, as some tree tools may be unavailable on certain platforms.
- Install script is required for AMD GPU support.
- Docker (built from the provided Dockerfile) is recommended for full support for iterative mode.
| Platform / Setup | Conda | Script | Docker |
|---|---|---|---|
| Linux (x86_64) | ✅ | ✅ | ✅ |
| Linux (aarch64) | ✅ | ✅ | 🟡 |
| macOS (Intel Chip) | ✅ | ✅ | ✅ |
| macOS (Apple Silicon) | ✅ | ✅ | 🟡 |
| NVIDIA GPU | ✅ | ✅ | ✅ |
| AMD GPU | ❌ | ✅ | ❌ |
Note
🟡 The Docker image is currently built for the linux/amd64 platform. While it can run on arm64 systems (e.g., Apple Silicon or Linux aarch64) via emulation, this may lead to reduced performance.
Note
⚠️ To enable GPU support, the appropriate GPU drivers must be installed on the host system. This applies to all installation methods (Installation script, Conda, and Docker). The CUDA toolkits and libraries are included in Conda and Docker setups, but must be installed manually when using the installation script.
Using Conda
TWILIGHT is available on multiple platforms via Conda. See TWILIGHT Bioconda Page for details.
- Install Conda (if not installed)
# Replace <PLATFORM> with the actual platform of your system # See https://repo.anaconda.com/miniconda/ for details wget https://repo.anaconda.com/miniconda/Miniconda3-latest-<PLATFORM>.sh chmod +x Miniconda3-latest-<PLATFORM>.sh ./Miniconda3-latest-<PLATFORM>.sh export PATH="$HOME/miniconda3/bin:$PATH" # Reload shell configuration source ~/.bashrc # For Bash (Most Linux) source ~/.zshrc # For Zsh (Default in Mac) - Create and activate a Conda environment
- Install TWILIGHT iterative mode
Using installation script (requires sudo access)
Users without sudo access are advised to install TWILIGHT via Conda or Docker.
- Clone the repository
- Install dependencies (requires sudo access)
TWILIGHT depends on the following common system libraries, which are typically pre-installed on most development environments: It also requireslibtbb-dev, which is not always pre-installed on all systems. For users who do not have sudo access and are missing onlylibtbb-dev, our script builds and installs TBB from source in the local user environment, with no sudo access required.
For Ubuntu users with sudo access, if any of the required libraries are missing, you can install them with: For Mac users, install dependencies using Homebrew: - Build TWILIGHT
Our build script automatically detects the best available compute backend (CPU, NVIDIA GPU, or AMD GPU) and builds TWILIGHT accordingly. Alternatively, users can manually specify the desired target platform.
Automatic build: Build for a specific platform: - The TWILIGHT executable is located in the
bindirectory and can be run as follows: - (optional) Install TWILIGHT iterative mode (ensure Conda is installed first)
Using Dockerfile
The Dockerfile installed all the dependencies and tools for TWILIGHT default/iterative mode.
- Clone the repository
- Build the docker image
CPU version
GPU version (using nvidia/cuda as base image)
- Start and run docker container
CPU version GPU version
- Run TWILIGHT
Run TWILIGHT
TWILIGHT Command Line Interface (CLI)
Main Functionalities
- Build MSA from unaligned Sequences (Default Mode)
- Merging Multiple MSAs
- Add New Sequences to an Existing Alignment
Command Line Options
| Option | Description |
|---|---|
| Inputs | |
| Build MSA from unaligned Sequences (Default Mode) | |
-t, --tree |
Input guide tree file in Newick format (required). |
-i, --sequences |
Input unaligned sequences file in FASTA format (required). |
| Merging Multiple MSAs | |
-f, --files |
Path to the directory containing all MSA files in FASTA format to be merged. |
| Add New Sequences to an Existing Alignment | |
-t, --tree |
Input tree file in Newick format with placements for sequences to be aligned (optional). |
-i, --sequences |
Input unaligned sequences file in FASTA format (required). |
-a, --alignment |
Backbone MSAs in FASTA format (required). |
| Outputs/Files | |
-o, --output |
Output MSA file name (required). |
-d, --temp-dir |
Directory for storing temporary files. Used when -f or -m is specified. |
-k, --keep-temp |
Keep the temporary directory (default: disabled). |
--overwrite |
Force overwriting the temporary and output files (default: disabled). |
--write-prune |
Write the pruned tree to the output directory (must be used with --prune, default: disabled). |
| Hardware Usage | |
-C, --cpu |
Number of CPU cores (default: all available cores). |
| Options only for GPU version | |
-G, --gpu |
Number of GPU (default: all available GPUs). |
--gpu-index |
Specify the GPU to be used, separated by commas. For example, 0,2,3 uses 3 GPUs with the index 0, 2 and 3. |
--cpu-only |
Run the program only using CPU. |
| Alignment Parameters | |
--type |
Data type: n for nucleotide, p for protein. Automatically inferred if not specified. |
-m, --max-subtree |
Maximum number of leaves per subtree; enables divide-and-conquer method (default: INT_MAX). |
-r, --remove-gappy |
Threshold for removing gappy columns (set to 1 to disable, default: 0.95). |
--prune |
Prune the input guide tree based on the presence of unaligned sequences (default: disabled). |
-w, --wildcard |
Treat unknown or ambiguous bases as wildcards and align them to usual letters (default: disabled). |
--no-align-gappy |
Do not align gappy columns; this will create a longer MSA (default: disabled). |
--length-deviation |
Sequences whose lengths deviate from the average by more than the specified fraction will be deferred or excluded (default: disabled). |
--max-ambig |
Sequences with an ambiguous character proportion exceeding the specified threshold will be deferred or excluded (default: 0.1). |
--filter |
Exclude sequences with high ambiguity or length deviation (default: disabled.) |
--merge |
Method to merge subtrees: t for Transitivity Merger and p for Progressive Alignment (default: p). |
| Scoring Parameters | |
--match |
Match score (default: 18). |
--mismatch |
Mismatch penalty for transversions (default: -8). |
--transition |
Score for transition (default: -4). |
--gap-open |
Gap-open penalty (default: -50). |
--gap-extend |
Gap-extend penalty (default: -5). |
-x, --matrix |
User-specified substitution matrix path. |
--xdrop |
X-drop value (scale). The actual X-drop will be multiplied by the gap-extend penalty (default: 600). |
| General | |
--check |
Check the final alignment. Sequences with no legal alignment will be displayed. |
-v, --verbose |
Print out every detail process. |
-h, --help |
Print help messages and exit. |
--version |
Show TWILIGHT version and exit. |
Usages and Examples
Note
All files used for the examples below can be found in TWILIGHT/dataset.
Enter into the build directory (assuming $TWILIGHT_HOME directs to the TWILIGHT repository directory)
Build MSA from unaligned Sequences (Default Mode)
Generate MSA by giving an unaligned sequence file and a guide tree file.
- Usage syntax
- Example
Merge Multiple MSAs
Assume a star-tree structure to merge multiple MSAs. Please move all MSA files to be merged into one folder.
- Usage syntax
- Example
Add New Sequences to an Existing Alignment
For better accuracy, it is recommended to use a tree that includes placements for the new sequences. If no tree is provided, TWILIGHT aligns new sequences to the profile of the entire backbone alignment, which may reduce accuracy. In this case, using the provided Snakemake workflow is advised.
- Usage syntax
- Example
Divide-and-Conquer Method
Divide tree into subtrees with at most m leaves and align them sequentially to reduce the CPU’s main memory usage.
- Usage syntax
- Example
Flexible Tree Support
Prunes tips that are not present in the unaligned sequence file. This is useful when working with a large tree but only aligning a subset of sequences, without needing to re-estimate the guide tree. Outputting the pruned tree is also supported.
- Usage syntax
- Example
Snakemake Workflow
TWILIGHT uses the Snakemake workflow to integrate external tools for estimating guide trees and placing new sequences. These are used in iterative mode or when aligning new sequences to existing alignments. For setting up the environment and installing external tools, see here.
Main Functionalities
Command Line Configurations
| Configurations | Description and Options |
|---|---|
| Snakemake Workflow Configurations | |
-n |
Dry run, check workflow. |
--cores |
Number of CPU cores to use (default: all available cores). Some Snakemake versions may require this to be explicitly set. |
User-defined Configurations (used with --config) |
|
| Iterative Mode | |
SEQ |
Input unaligned sequences file in FASTA format (required). |
INITTREE |
Tree method for initial guide tree, options: parttree, maffttree or mashtree. (default: parttree). |
ITERTREE |
Tree method for for intermediate steps, options: rapidnj or fasttree. (default: rapidnj). |
| Add New Sequences to an Existing Alignment (Placement Mode) | |
SEQ |
New sequences in FASTA format to be placed and aligned (required). |
ALN |
Backbone alignment in FASTA format for placing new sequences (required.) |
TREE |
Backbone tree (optional; FastTree will be used for estimation if not provided). |
| General | |
TYPE |
Input sequence type, options: n for nucleotide or p for protein sequences (required). |
OUT |
Output MSA file path (required). |
DIR |
Directory for storing temporary files. |
ITER |
Number of iterations (default: 3 for Iterative Mode and 2 for Placement Mode). |
FINALTREE |
Final tree estimation method (skip if unspecified), options: fasttree, raxml or iqtree. |
KEEP |
Keep the temporary files, options: yes or no (default: no). |
OVERWRITE |
Overwrite the existing files, options: yes or no (default: no). |
Note
Default options in workflow/config.yaml will be used if the configuration is not specified. You can also modify workflow/config.yaml to set your options.
Note
For users who install TWILIGHT via Conda, please replace the executable path "../bin/twilight" with "twilight" in config.yaml. Feel free to switch to a more powerful tree tool if available, such as replacing "raxmlHPC" with "raxmlHPC-PTHREADS-AVX2" for better performance.
Usages and Examples
Enter workflow directory and type snakemake to view the help messages.
cd workflow
snakemake
# or, for Snakemake versions that require specifying total number of cores:
snakemake --cores 1
Iterative Mode
TWILIGHT iterative mode estimate guide trees using external tools.
- Usage syntax
- Example - Using default configurations
- Example - Generates the final tree based on the completed MSA.
Add New Sequences to an Existing Alignment (Placement Mode)
TWILIGHT aligns new sequences to the profile of the backbone alignment, infers their placement with external tools, and then refines the alignment using the inferred tree.
- Usage syntax
- Example - The backbone alignment is accompanied by a tree.
- Example - The backbone tree is unavailable, estimate it using external tools and generate a final tree after alignment.
Contributions
We welcome contributions from the community to enhance the capabilities of TWILIGHT. If you encounter any issues or have suggestions for improvement, please open an issue on TWILIGHT GitHub page. For general inquiries and support, reach out to our team.
Citing TWILIGHT
If you use the TWILIGHT in your research or publications, we kindly request that you cite the following paper:
Yu-Hsiang Tseng, Sumit Walia, Yatish Turakhia, "Ultrafast and ultralarge multiple sequence alignments using TWILIGHT", Bioinformatics, Volume 41, Issue Supplement_1, July 2025, Pages i332–i341, doi: 10.1093/bioinformatics/btaf212