Using RIVET for Other Pathogens
Below are two examples of using RIVET's backend pipeline to infer and visualize recombinants of other pathogens beyond SARS-CoV-2.
Warning
Currently, RIVET's backend QC/filtration pipeline is specific to SARS-CoV-2 and will not run when using the RIVET backend for other pathogens.
Human Respiratory Syncytial Virus (HRSV) Subgroup A
Below are the steps followed to infer putative recombinants in an RSV mutation-annotated tree (MAT).
Using RIVET frontend for Visualization
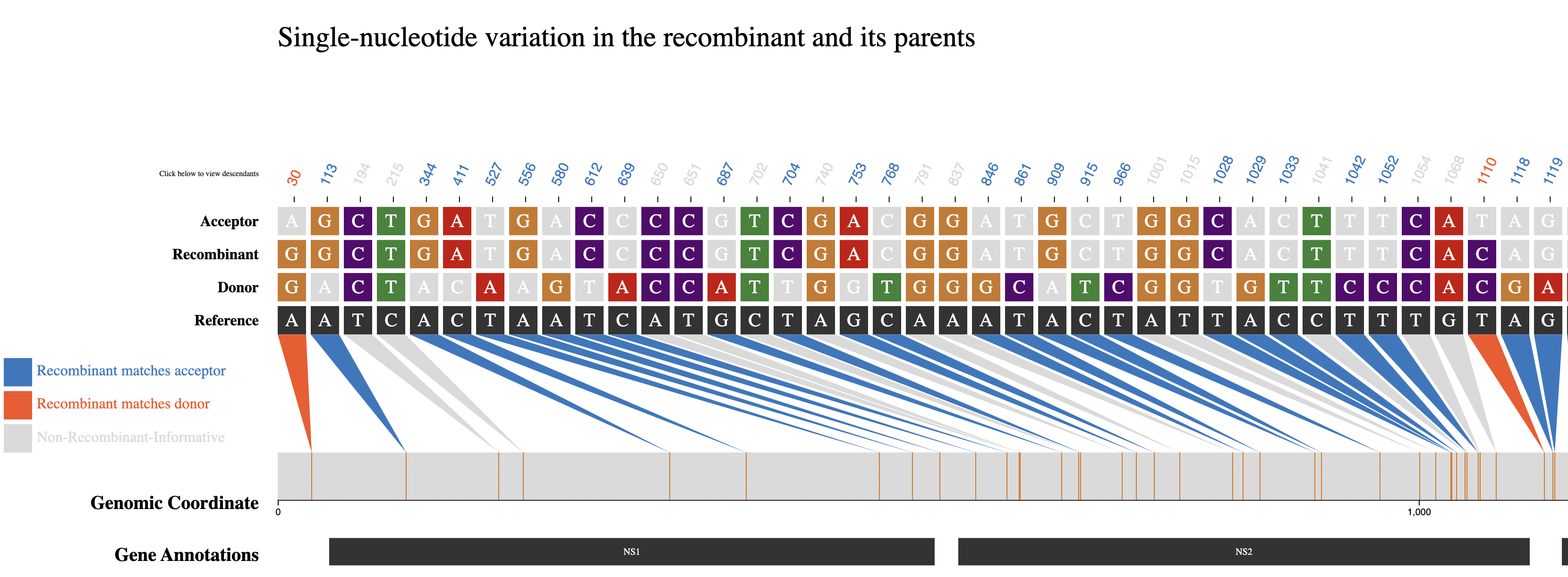
Since the SNV plot for RSV includes many sites, only the region up to around position 1000 is shown in the image above.
Please click the download button below to view the entire RSV SNV plot as an SVG image.
Monkeypox Virus
Edit the following fields in the config.yaml file:
Change the GenBank file from the default SARS-CoV-2 file to the corresponding GenBank file for your pathogen of interest, Monkeypox virus in this case.
Warning
Make sure the environment field is set to local.
port at which RIVET will host the local HTTP server in your browser.
Now run the following command and RIVET will automatically open your browser to launch the frontend results table and SNV visualization.
python3 rivet-frontend.py -r recombination_mpxv.2023-07-01.tsv -v mpxv.2023-07-01.vcf -c config.yaml
Below is the SNV plot we get for one of the monkeypox virus inferred recombinants.
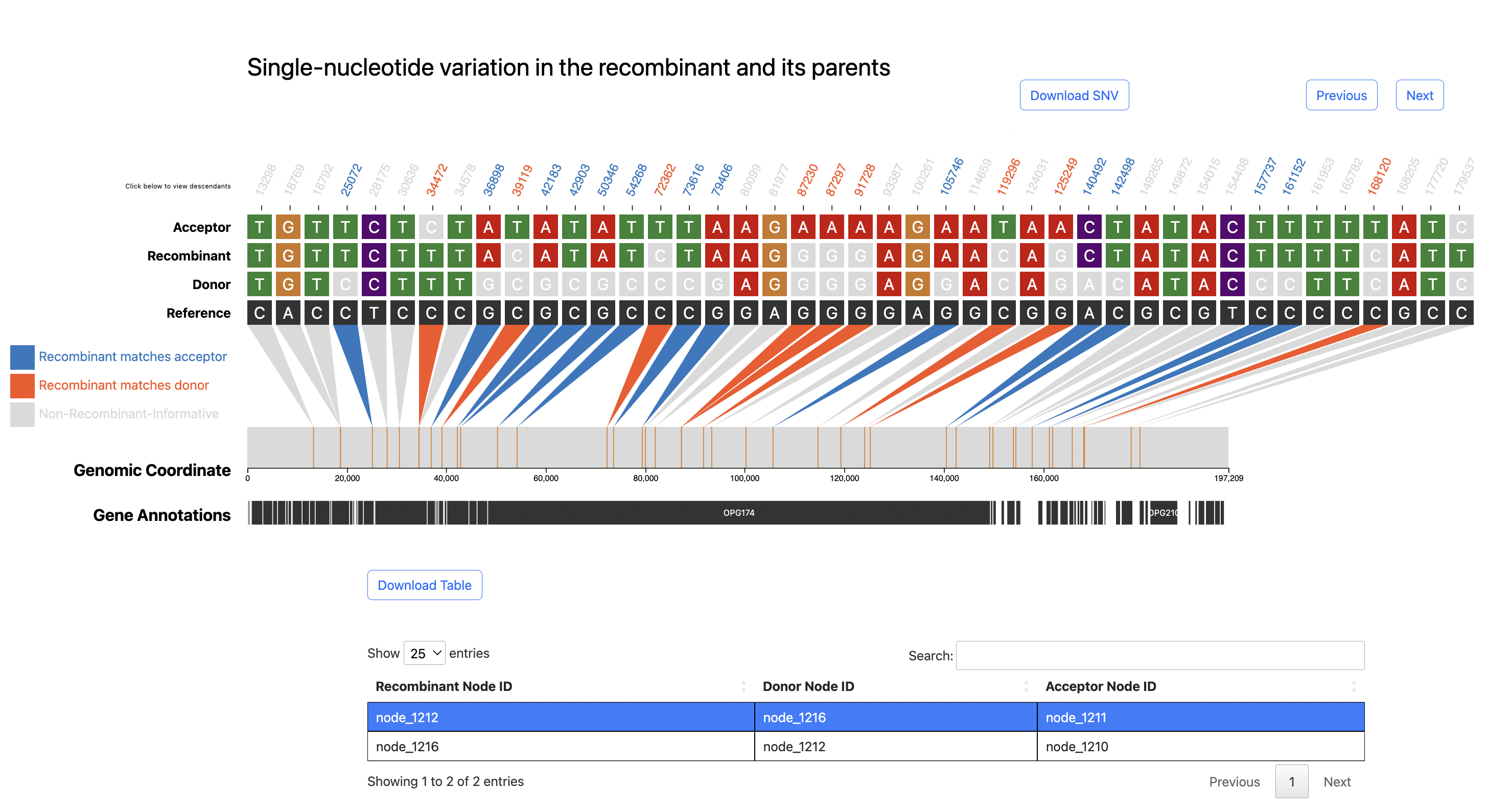
Check
For pathogens with larger genomes than SARS-CoV-2, you may want to change the step interval of genomic coordinate tick marks. This can be done by changing the tick_step field in RIVET frontend config.yaml file.